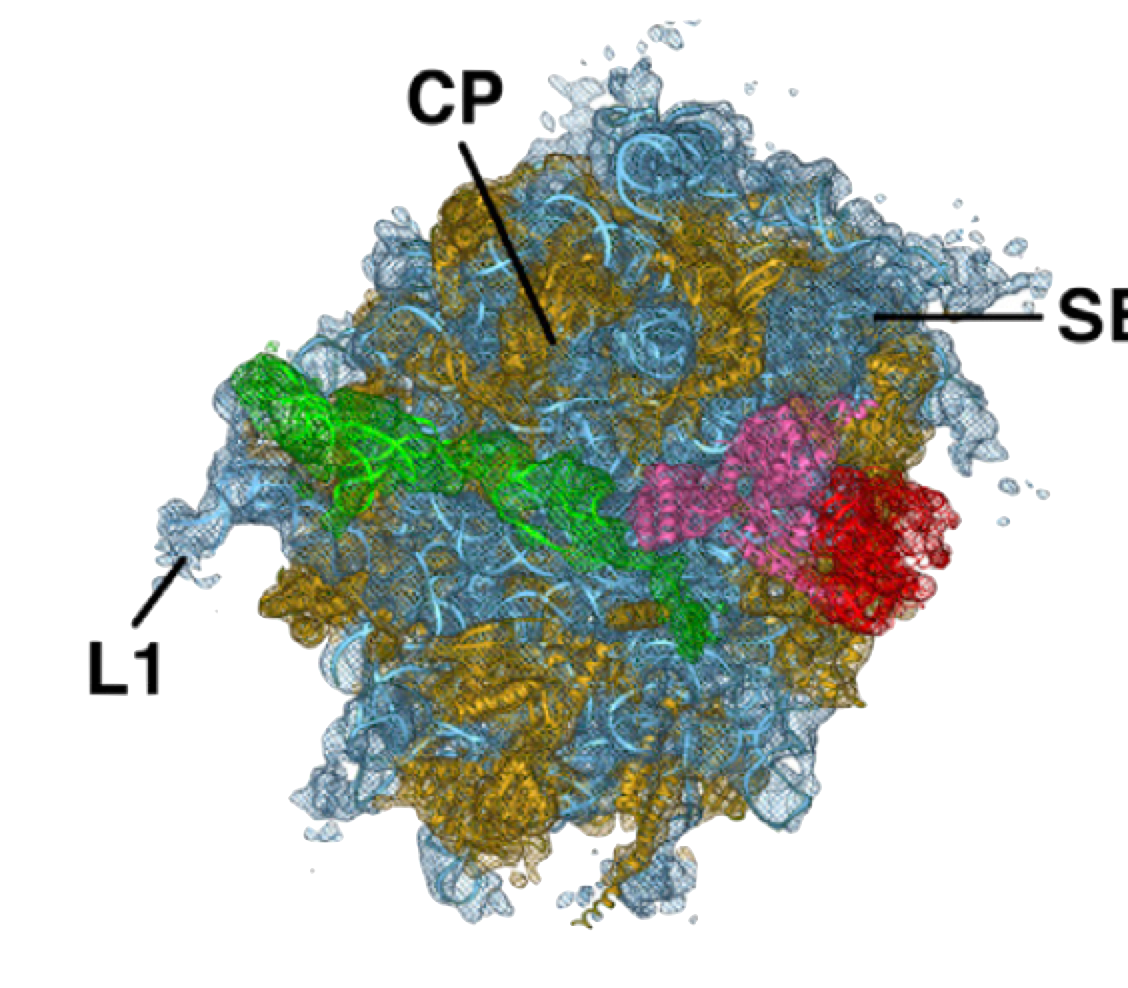
PDB 4D5Y | EMDB 2810
Cryo-EM of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus IRES
Muhs et al. (2015) Cryo-EM of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus IRES. Molecular Cell 57 (3), 422-432, published February 5, 2015. doi:10.1016/j.molcel.2014.12.016. View PDF »
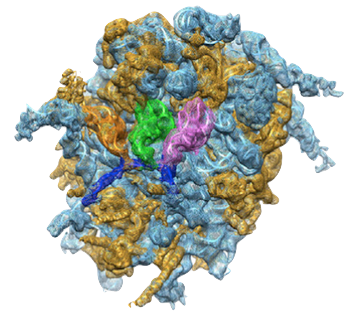
PDB 4UJE | EMDB 2620
Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement
Budkevich et al. (2014) Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement. Cell 158 (1), 121-131, published July 3, 2014. doi:10.1016/j.cell.2014.04.044. View PDF »
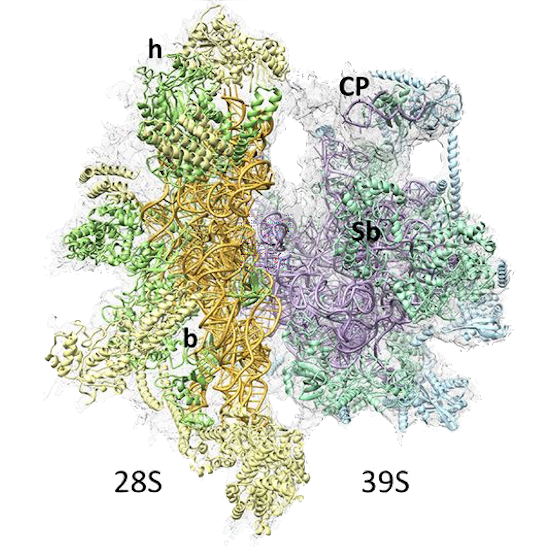
PDB 3JD5 | EMDB 5941
Cryo-EM Structure of the Small Subunit of the Mammalian Mitochondrial Ribosome
Kaushal et al. (2014) Cryo-EM Structure of the Small Subunit of the Mammalian Mitochondrial Ribosome. Proceedings of the National Academy of Sciences 111 (20), 7284-7289, published May 20, 2014. doi:10.1073/pnas.1401657111. View PDF »
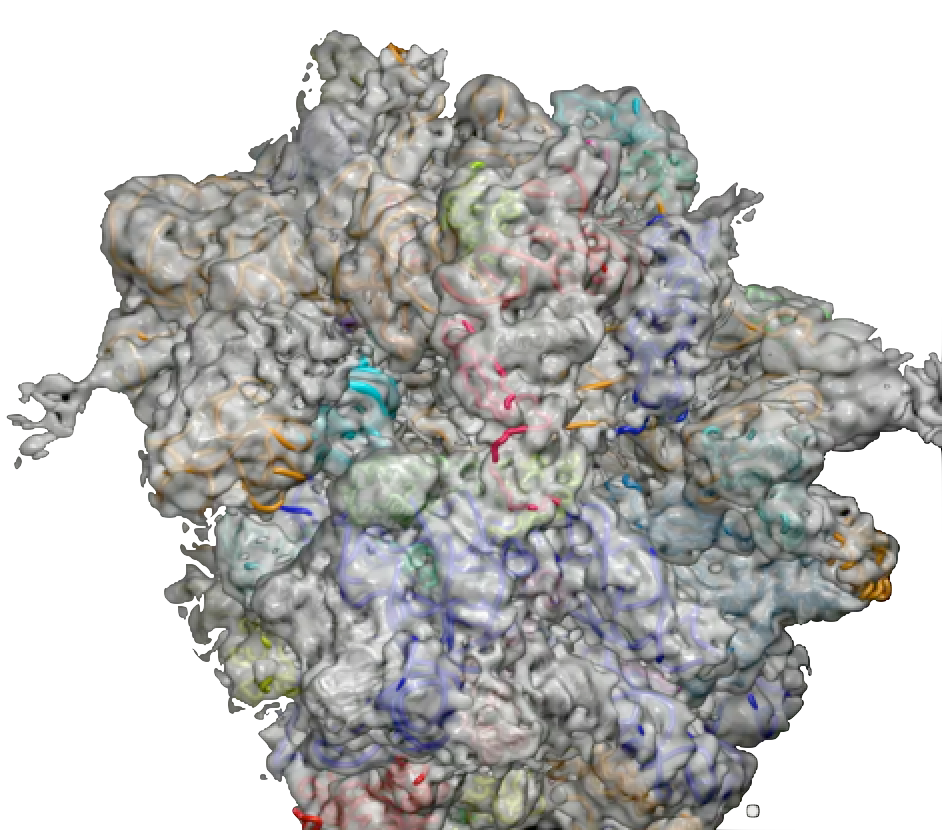
PDB 4V5N | EMDB 1799
Head Swivel on the Ribosome Facilitates Translocation Via Intra-Subunit tRNA Hybrid Sites
Ratje et al. (2010) Head Swivel on the Ribosome Facilitates Translocation Via Intra-Subunit tRNA Hybrid Sites. Nature 468 (7324), 713-716, published December 2, 2010. doi:10.1038/nature09547. View PDF »
All-Atom Homology Model of the Escherichia coli 30S Ribosomal Subunit
Tung et al. (2002) All-Atom Homology Model of the Escherichia coli 30S Ribosomal Subunit. Nature Structural & Molecular Biology 9 (10), 750-755, published September 16, 2002. doi:10.1038/nsb841. View PDF »